Breaking Barriers in Genomics: Slorado Brings Open-Source Genomic Basecalling to AMD GPUs
Nov 24, 2025

UNSW Sydney, Pawsey, and AMD Collaborate to Democratize DNA Analysis
DNA sequencing is entering a new era of speed and openness. Researchers at UNSW Sydney and AMD Research and Advanced Development (RAD) have jointly developed Slorado, the world’s first fully open-source tool for real-time nanopore DNA decoding. Built on AMD GPUs and the ROCm™ open-source platform, Slorado delivers high-performance, scalable basecalling to the genomics community, enabling scientists worldwide to analyze nanopore sequencing data on any GPU hardware. Slorado’s performance and scalability were demonstrated on the Setonix supercomputer at the Pawsey Supercomputing Research Centre, Australia.
Accelerating Genomics with Open Compute
Nanopore sequencing is transforming genomics with affordable, portable, and real-time DNA analysis. Devices from Oxford Nanopore Technologies (ONT) generate continuous electrical signals, or squiggles, that an AI model decodes into DNA bases (A, C, G, T). This decoding process is known as basecalling. ONT’s open-source Dorado basecaller has driven significant advances in this space, but until now, it has relied on closed-source NVIDIA components, limiting researchers to specific GPUs.
Slorado removes this barrier for researchers by offering a fully open-source alternative that supports the latest transformer-based basecalling models and executes efficiently on AMD Instinct™ and Radeon™ GPUs through ROCm. By delivering full transparency, portability, and performance, Slorado gives researchers the freedom to deploy nanopore workflows across diverse computing environments, from supercomputers to local GPU workstations.
Demonstrated on Australia’s Flagship AMD-Powered Supercomputer
The Setonix supercomputer at the Pawsey Supercomputing Research Centre, powered by AMD Instinct MI250X GPUs, served as the primary testbed for Slorado’s performance validation.

A typical nanopore sequencing run of a human genome generates approximately 1 terabyte of raw signal data over 48 hours. By leveraging multi-node scaling on Setonix, researchers can process these datasets in parallel, dramatically accelerating time-to-insight and enabling large-scale genomic studies. Slorado processed a human genome dataset in as little as 2.3 hours, dramatically faster than traditional workflows.
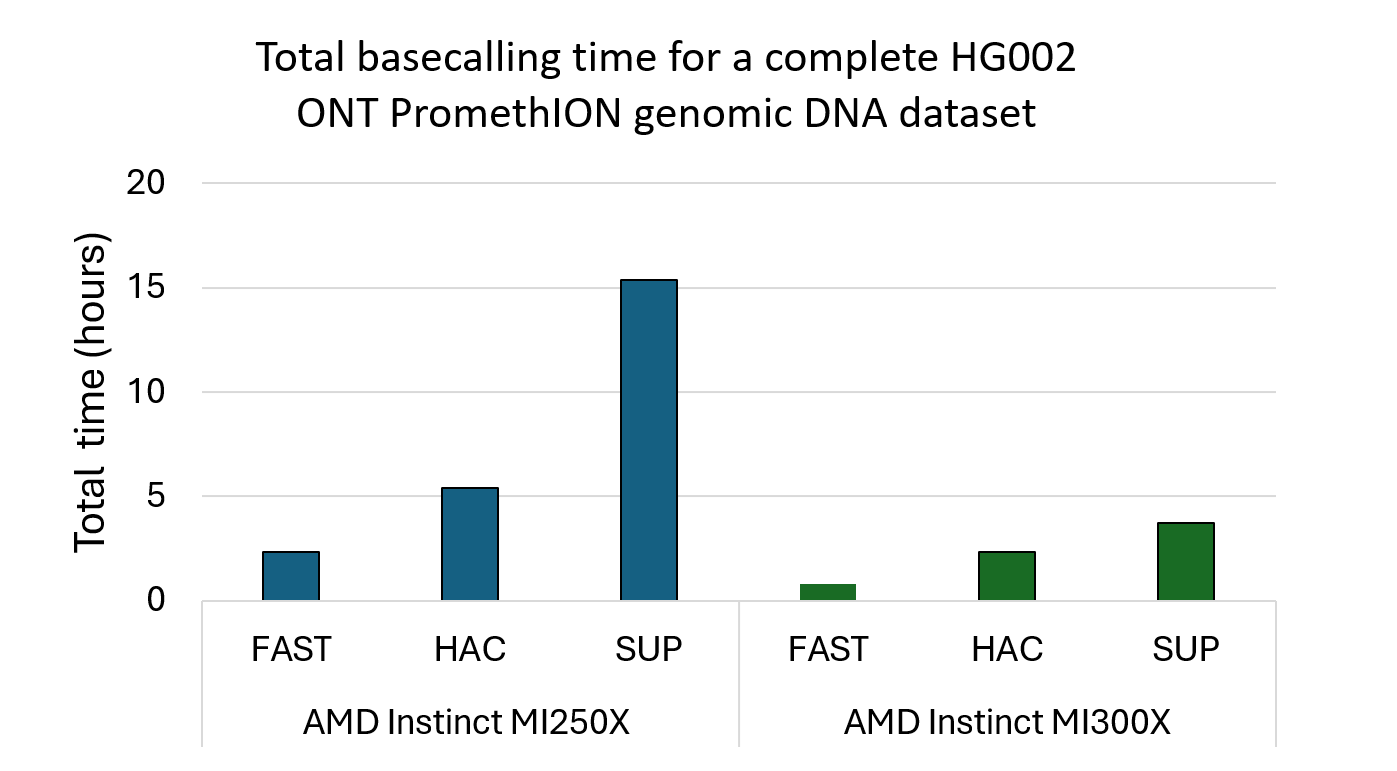
Figure 1 compares total basecalling time across AMD Instinct MI250X and AMD Instinct MI300X GPUs for the FAST, HAC, and SUP v5 models. On Pawsey’s Setonix system, which is powered by four AMD Instinct MI250X accelerators per node, Slorado achieved total basecalling times of 2.3 hours for the FAST model, 5.4 hours for the HAC model, and 15.4 hours for the computationally intensive SUP model when processing a complete HG002 ONT PromethION dataset. These results demonstrate strong throughput and efficient scaling on the MI250X architecture currently deployed in large HPC environments. Slorado was also evaluated on an eight-way configuration of AMD Instinct MI300X, a more recent accelerator generation designed to support higher memory capacity and AI-centric workloads. On MI300X, the same dataset completed in 0.8 hours for FAST, 2.3 hours for HAC, and 3.7 hours for SUP, illustrating the significant performance uplift available on this generation and the efficiency gains Slorado achieves when paired with higher-bandwidth, higher-capacity GPU architectures.
This work was led by Dr. Hasindu Gamaarachchi, together with his PhD student, Bonson Wong from UNSW Sydney.
Basecalling on AMD GPUs – Quick Start Guide
The easiest way to get started is by using the precompiled Slorado binaries for Linux.
Step 1: Download the latest x86_64-rocm-linux binaries
VERSION=v0.3.0-beta
wget "https://github.com/BonsonW/slorado/releases/download/$VERSION/slorado-$VERSION-x86_64-rocm-linux-binaries.tar.xz"
Step 2: Extract the tarball and verify installation
tar xvf slorado-$VERSION-x86_64-rocm-linux-binaries.tar.xz
cd slorado-$VERSION
bin/slorado --help
Step 3: Download the test dataset (20,000 reads)
wget -O PGXXXX230339_reads_20k.blow5 https://slow5.bioinf.science/hg2_prom_5khz_subsubsample
Step 4: Run the basecaller with a model of your choice
Example using the v5.0.0 High Accuracy model:
./bin/slorado basecaller models/dna_r10.4.1_e8.2_400bps_hac@v5.0.0 PGXXXX230339_reads_20k.blow5 -o out.fastq
If the binaries are incompatible with your system, Slorado can be compiled from source following the instructions at:
Open Science. Open Compute. Open Future for Research.
Slorado demonstrates how open science and open computing can work together to advance discovery. By integrating cutting-edge AI-based basecalling methods with the open ROCm software stack, AMD and its collaborators are empowering researchers to push the boundaries of genomic analysis without being constrained by proprietary platforms.
Through initiatives like Slorado, AMD continues to enable a new generation of scientific computing that is scalable, transparent, and accessible to all.
- Read the Pawsey blog: Pawsey enables more flexible and scalable DNA analysis
- Dive into the technical details: Nanopore basecalling on Pawsey supercomputer using AMD GPUs
- Watch the live demo and Q&A: YouTube Recording
Disclaimer
Slorado incorporates components licensed under the Oxford Nanopore Technologies PLC Public License v1.0. Use is permitted for research purposes only.